2026.01.16 Friday
Exploring Mutations That Spontaneously Switch on a Key Brain Cell Receptor
Researchers uncover insights that pave the way for a better understanding of brain disorders and targeted drugs
Many people are familiar with histamine, a biological molecule, that serves as a key driver of allergic reactions and other immune responses. However, histamine is also a major neurotransmitter in the mammalian brain, regulating essential cognitive functions like wakefulness, attention, and learning. Histamine levels are partially kept in check by the histamine H3 receptor (H3R), belonging to the G protein-coupled receptor (GPCR) family. In essence, H3R acts as a ‘braking system’ in the central nervous system, modulating the release of histamine and various neurotransmitters to maintain proper balance. Because of this gatekeeping role, optimized H3R activity is vital for health, and problems in this signaling pathway have been linked to neurological conditions like attention-deficit hyperactivity disorder, schizophrenia, and Alzheimer’s disease.
Despite its importance, studying the intricacies and functionalities of H3R is challenging. Scientists often use simplified laboratory models such as yeast to study GPCRs because they are easier to genetically manipulate and analyze. However, H3R becomes ‘silent’ or inactive when expressed in yeast, creating a significant hurdle for research. Furthermore, a major knowledge gap persists regarding what’s known as a ‘constitutive activity’ in GPCRs--the ability of a receptor to spontaneously turn to an ‘active’ state even in the absence of ligand (trigger) molecules. While certain genetic mutations can induce this phenomenon, the exact molecular mechanisms underlying the necessary changes remain unclear.
To address these questions, a research team led by Professor Mitsunori Shiroishi from the Department of Biological Science and Technology, Tokyo University of Science (TUS), Japan, investigated the structural features that control how H3R turns signaling on and off. The study was made available online on December 22, 2025, and will be published in Volume 35, Issue 1 of the journal Protein Science on January 01, 2026. The study was co-authored by Ms. Ami Nakajima, who completed her Master’s course in 2024, as well as first-year doctoral student Mr. Hiroto Kaneko and Assistant Professor Kosuke Oyama, all from TUS. Together, the researchers explored how specific amino acid mutations in H3R affect the receptor’s constitutive activity, measuring the physicochemical changes these mutations induced. They focused on four specific point mutations (L732.43M, F193ECL2S, S3596.36Y, and C4157.56R) that were previously identified as capable of restoring H3R activity in yeast.
The team employed a multifaceted approach to observe these mutations in action. They created double mutants to see if the changes acted cooperatively and tested the variants in both yeast and mammalian cell models. They also used radioligand binding assays to measure the mutant receptors’ affinity to histamine and specialized chromatography techniques to gauge their structural stability. Finally, they introduced the same mutations into the related histamine H1 receptor (H1R) to determine if the observed effects were consistent across different histamine receptor types.
These experiments revealed a striking link between activity and stability. Simply put, these mutations not only increased constitutive activity by pushing H3R into a spontaneous ‘ON’ state, but also compromised the receptor’s physical integrity. The team’s analysis showed that the most active mutants were also the most unstable. Interestingly, these mutations did not significantly change how well histamine binds to the receptor, suggesting that they purely influence the internal ‘switch’ mechanism rather than the external docking site.
The team also revealed that these effects are mostly receptor-specific. When the team applied the same mutations to H1R, only one mutation (C4157.56R) showed a similar enhancement in activity. This highlights that even closely related receptors have unique internal environments that dictate how they respond to structural changes. “Collectively, our findings reveal a close relationship between structural destabilization and constitutive activation in H3R, while also underscoring the complexity of GPCR activation,” remarks Prof. Shiroishi.
Considering that GPCRs are prominent drug targets for a wide variety of conditions, understanding how mutations affect their activity and stability is an essential knowledge for drug development. Moreover, since abnormal changes in receptor activity are often associated with pathological conditions, activating or deactivating these proteins as needed could help treat complex disorders. “We believe our findings will provide a foundation for a better understanding of brain diseases and the design of safer drugs, as well as artificial GPCRs for therapeutic and biotechnological applications,” concludes Prof. Shiroishi. The insights gained through this work may eventually lead to the creation of custom-engineered receptors in neuroscience to control specific brain circuits or the development of highly selective drugs that can fine-tune H3R activity with unprecedented precision.
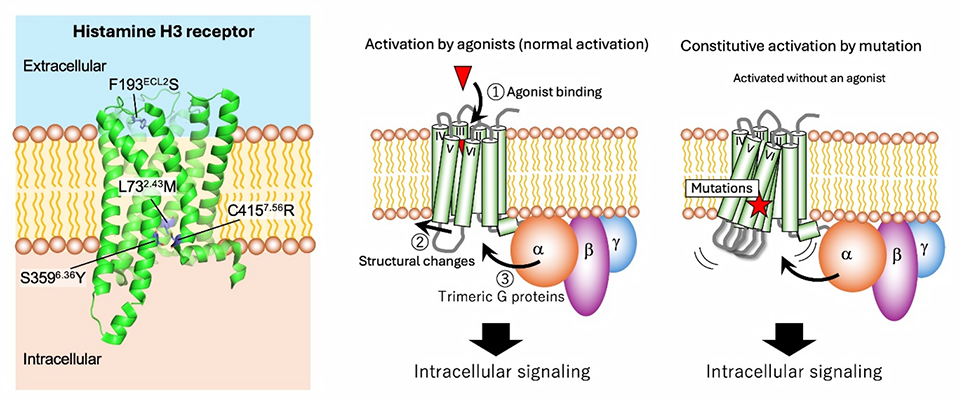
Image title: Activation of the histamine H3 receptor
Image caption: The histamine H3 receptor typically becomes active upon binding to one of its agonists, such as histamine itself. This, in turn, gives way to intracellular interactions with compounds known as trimeric G proteins, triggering downstream signaling effects. However, some mutations in the histamine H3 receptor can render it continuously active, even in the absence of an agonist. Understanding how these mutations affect receptor activity, and their implications in disease, is an important area of study within medicine and biochemistry.
Image credit: Professor Mitsunori Shiroishi from Tokyo University of Science, Japan
License type: Original content
Usage restrictions: Cannot be reused.
Reference
| Title of original paper | : | Activity-restoring mutations in the histamine H3 receptor increase constitutive activity and reduce structural stability |
| Journal | : | Protein Science |
| DOI | : | 10.1002/pro.70408 |
About The Tokyo University of Science
Tokyo University of Science (TUS) is a well-known and respected university, and the largest science-specialized private research university in Japan, with four campuses in central Tokyo and its suburbs and in Hokkaido. Established in 1881, the university has continually contributed to Japan's development in science through inculcating the love for science in researchers, technicians, and educators.
With a mission of "Creating science and technology for the harmonious development of nature, human beings, and society," TUS has undertaken a wide range of research from basic to applied science. TUS has embraced a multidisciplinary approach to research and undertaken intensive study in some of today's most vital fields. TUS is a meritocracy where the best in science is recognized and nurtured. It is the only private university in Japan that has produced a Nobel Prize winner and the only private university in Asia to produce Nobel Prize winners within the natural sciences field.
■
Tokyo University of Science(About TUS)

About Professor Mitsunori Shiroishi
from Tokyo University of Science
Dr. Mitsunori Shiroishi graduated from the Department of Biomolecular Engineering at Tohoku University, Japan, in 1998 and completed his doctoral degree there in 2003. He joined Tokyo University of Science in April 2018, where he currently serves as a full-time Professor. He leads the Protein Engineering Laboratory as Principal Investigator, where his team conducts cutting-edge research on protein science focused mainly on cell receptors with pharmaceutical potential, including receptors with immunoglobin-like fold and G protein-coupled receptors. He has published over 70 research papers on these topics.
About Assistant Professor Kosuke Oyama
from Tokyo University of Science
Funding information
This work was funded in part by JSPS KAKENHI Grant Numbers JP25709080 and JP15K14460; the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from MEXT and AMED, and the Kato Memorial Bioscience Foundation.

