2025.11.04 Tuesday
Advanced Molecular Dynamics Simulations Capture RNA Folding with High Accuracy
Researchers test state-of-the-art models on diverse RNA structures, opening doors for RNA-based therapies and drug design
Ribonucleic acid (RNA) is one of life's most versatile molecules, with roles going far beyond being a messenger of genetic code, as it is fundamentally involved in gene regulation, processing, and maintenance across all living systems. This versatility is deeply tied to RNA's ability to adopt complex three-dimensional shapes, known as secondary and tertiary structures. With the global rise of RNA-based therapeutics, understanding and precisely predicting secondary and tertiary structures is essential to fully harness the RNA's potential in biotechnology.
Despite the need to model these structures, simulating the complete folding process of an RNA molecule from an initially unfolded state remains a challenge in computational biophysics. These molecular dynamics (MD) simulations involve many physics-based computational potentials (or 'force fields') and special methods to adjust to different folding timescales. Historically, success in this area has been limited. Although many different modeling approaches have been tested, prior MD simulation studies only reported the accurate folding of a few, simple stem-loop structures.
Against this backdrop, Associate Professor Tadashi Ando from the Department of Applied Electronics, Tokyo University of Science, Japan, conducted a large-scale, rigorous evaluation of modern simulation methods. In his latest paper published in the journal ACS Omega on October 26, 2025, he explored whether a combination of state-of-the-art computational tools could reliably model the fundamental folding process of a significantly larger and more structurally diverse library of RNA stem loops than ever tested before.
The study employed conventional MD simulations using two cutting-edge computational components: the DESRES-RNA atomistic force field (a potential function refined for highly accurate RNA simulations) and the GB-neck2 implicit solvent model. This implicit solvent model is the key, as it approximates the surrounding liquid as a continuous medium instead of actual molecules. This dramatically accelerated the rate of conformational sampling compared to explicit solvent models, which are much more computationally demanding. Dr. Ando applied this methodology to a varied set of 26 RNA stem-loops, ranging from 10 to 36 nucleotides in length and structures featuring bulges and internal loops. Worth noting, all simulations started from fully extended, unfolded conformations.
The results demonstrated a high degree of folding stability and accuracy, validating the predictive power of the combined DESRES-RNA and GB-neck2 tools. A total of 23 out of 26 RNA molecules successfully folded into the expected structures. For the 18 simpler stem loops, this folding was achieved with exceptional accuracy, showing root mean square deviation (RMSD) values--a measure of deviation from the known experimental structure--of less than 2 Å for the stem and less than 5 Å for the molecules.
Even among the more challenging motifs, five of the eight containing bulges or internal loops were successfully achieved. For these complex structures, the simulations also revealed a distinct folding pathway. "Previous studies have reported only two or three folding examples of simple stem-loop structures of approximately 10 residues, whereas this study deals with structures of much greater scale and complexity," highlights Dr. Ando.
The successful simulation of folding across such a broad and complex library represents a significant milestone in computational biology. It rigorously validates the chosen combination of force field and solvent models as a reliable starting point for investigating large-scale conformational changes in RNA. The study also unveiled directions for future refinement of these tools. While stem regions were highly accurate, the loop regions showed less accuracy, with RMSD values of approximately 4 Å. This indicates that the implicit solvent model parameters need optimization, particularly for modeling non-canonical base pairs and incorporating the critical effects of divalent cations like magnesium.
Overall, this study represents an essential step to improve available RNA-based medical tools. Understanding how RNA folds is crucial when designing drugs that can target it, which could lead to new treatments for genetic disorders, viral infections such as COVID-19 or influenza, and certain cancers.
"The ability to reproduce the overall folding of the basic stem-loop structure is an important milestone in understanding and predicting the structure, dynamics, and function of RNA using highly accurate and reliable computational models," concludes Dr. Ando. "I expect this computational technique to lead to applications in RNA molecule design and drug discovery."
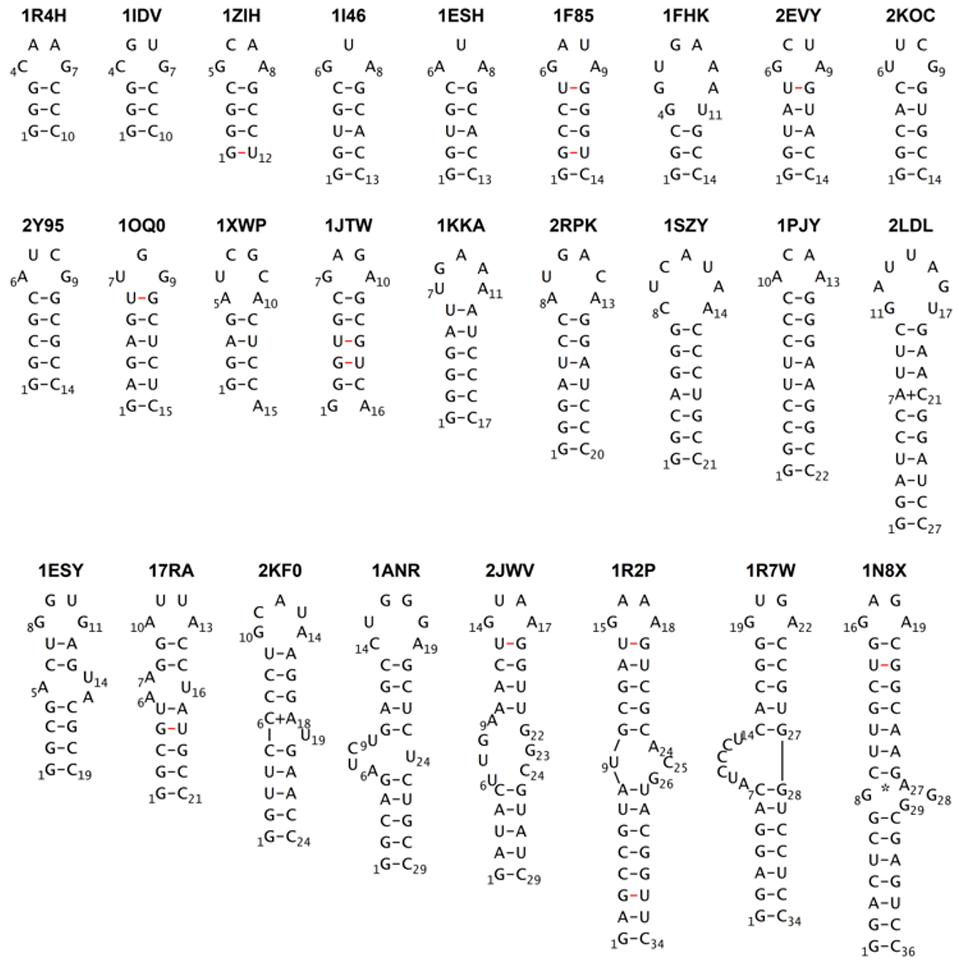
Image title: Library of RNA stem loops for molecular dynamics simulations
Image caption: This image depicts the base pairs and simplified structure of the stem loops simulated in this study.
Image credit: Associate Professor Tadashi Ando from Tokyo University of Science, Japan
License type: Original content
Usage restrictions: Cannot be reused without permission.
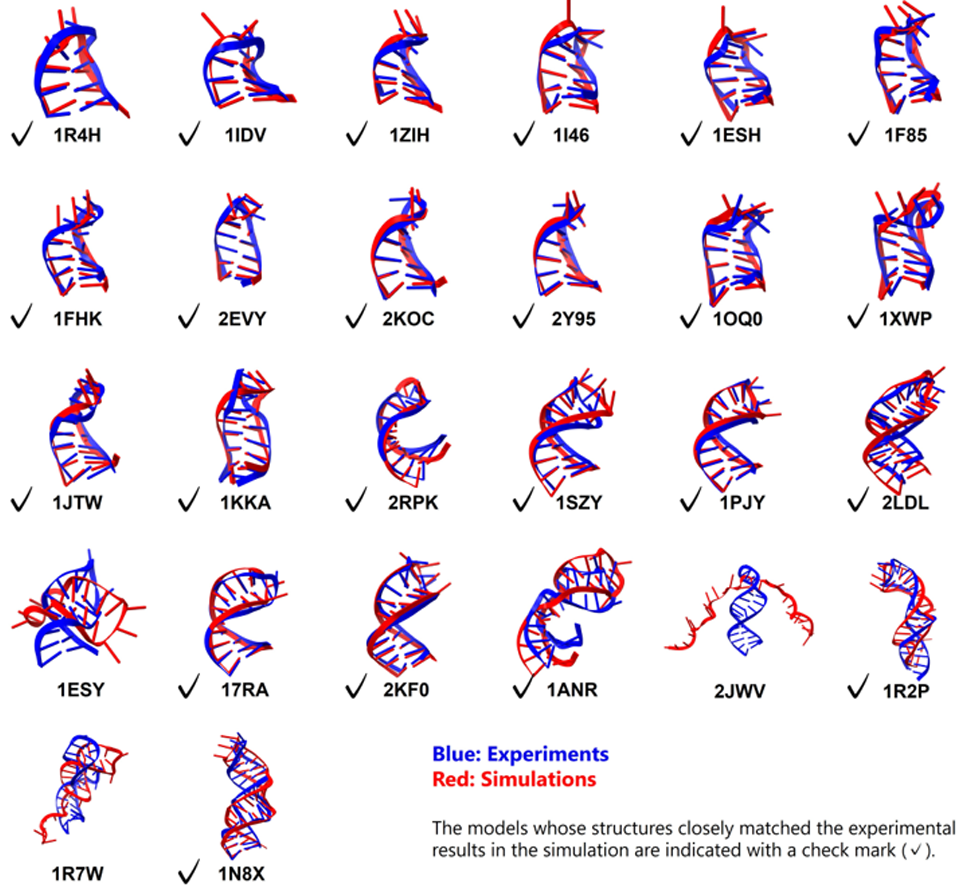
Image title: Structural comparison of RNA stem loops: simulation vs. experiment
Image caption: These images draw a comparison between the simulated folding and experimentally determined structures of RNA stem loops.
Image credit: Associate Professor Tadashi Ando from Tokyo University of Science, Japan
License type: Original content
Usage restrictions: Cannot be reused without permission.
Video title: Simulation of RNA stem-loop folding
Video caption: This video shows how molecular dynamics simulations can accurately recreate the RNA folding process.
Video credit: Associate Professor Tadashi Ando from Tokyo University of Science, Japan
License type: Original content
Usage restrictions: Cannot be reused without permission.
Reference
| Title of original paper | : | Molecular Dynamics Simulations of RNA Stem-Loop Folding Using an Atomistic Force Field and a Generalized Born Implicit Solvent |
| Journal | : | ACS Omega |
| DOI | : | 10.1021/acsomega.5c05377 |
About The Tokyo University of Science
Tokyo University of Science (TUS) is a well-known and respected university, and the largest science-specialized private research university in Japan, with four campuses in central Tokyo and its suburbs and in Hokkaido. Established in 1881, the university has continually contributed to Japan's development in science through inculcating the love for science in researchers, technicians, and educators.
With a mission of "Creating science and technology for the harmonious development of nature, human beings, and society," TUS has undertaken a wide range of research from basic to applied science. TUS has embraced a multidisciplinary approach to research and undertaken intensive study in some of today's most vital fields. TUS is a meritocracy where the best in science is recognized and nurtured. It is the only private university in Japan that has produced a Nobel Prize winner and the only private university in Asia to produce Nobel Prize winners within the natural sciences field.
■
Tokyo University of Science(About TUS)

About Associate Professor Tadashi Ando
from Tokyo University of Science
Dr. Tadashi Ando obtained his PhD degree from the Division of Engineering Science at Tokyo University of Science (TUS) Graduate School, Japan, in 2001. He currently serves as an Associate Professor at the Department of Applied Electronics of the Faculty of Advanced Engineering at TUS. He specializes in molecular simulations of biomolecules. He has published over 45 scientific articles on these topics.
Funding information
This research was financially supported by Tokyo University of Science.

