2024.02.20 Tuesday
Accelerating the Discovery of Single-Molecule Magnets with Deep Learning
The method identifies magnetic materials solely based on their crystal structure, eliminating the need for time-consuming experiments and simulations

Synthesizing or studying certain materials in a laboratory setting often poses challenges due to safety concerns, impractical experimental conditions, or cost constraints. In response, scientists are increasingly turning to deep learning methods which involve developing and training machine learning models to recognize patterns and relationships in data that include information about material properties, compositions, and behaviors. Using deep learning, scientists can quickly make predictions about material properties based on the material's composition, structure, and other relevant features, identify potential candidates for further investigation, and optimize synthesis conditions.
Now, in a study published on 1 February 2024 in the International Union of Crystallography Journal (IUCrJ), Professor Takashiro Akitsu, Assistant Professor Daisuke Nakane, and Mr. Yuji Takiguchi from Tokyo University of Science (TUS) have used deep learning to predict single-molecule magnets (SMMs) from a pool of 20,000 metal complexes. This innovative strategy streamlines the material discovery process by minimizing the need for lengthy experiments.
Single-molecule magnets (SMMs) are metal complexes that demonstrate magnetic relaxation behavior at the individual molecule level, where magnetic moments undergo changes or relaxation over time. These materials have potential applications in the development of high-density memory, quantum molecular spintronic devices, and quantum computing devices. SMMs are characterized by having a high effective energy barrier (Ueff) for the magnetic moment to flip. However, these values are typically in the range of tens to hundreds of Kelvins, making SMMs challenging to synthesize.
The researchers used deep-learning to identify the relationship between molecular structures and SMM behavior in metal complexes with salen-type ligands. These metal complexes were chosen as they can be easily synthesized by complexing aldehydes and amines with various 3d and 4f metals. For the dataset, the researchers worked extensively to screen 800 papers from 2011 to 2021, collecting information on the crystal structure and determining if these complexes exhibited SMM behavior. Additionally, they obtained 3D structural details of the molecules from the Cambridge Structural Database.
The molecular structure of the complexes was represented using voxels or 3D pixels, where each element was assigned a unique RGB value. Subsequently, these voxel representations served as input to a 3D Convolutional Neural Network model based on the ResNet architecture. This model was specifically designed to classify molecules as either SMMs or non-SMMs by analyzing their 3D molecular images.
When the model was trained on a dataset of crystal structures of metal complexes containing salen type complexes, it achieved a 70% accuracy rate in distinguishing between the two categories. When the model was tested on 20,000 crystal structures of metal complexes containing Schiff bases, it successfully discovered the metal complexes reported as single-molecule magnets. "This is the first report of deep learning on the molecular structures of SMMs," says Prof. Akitsu.
Many of the predicted SMM structures involved multinuclear dysprosium complexes, known for their high Ueff values. While this method simplifies the SMM discovery process, it is important to note that the model's predictions are solely based on training data and do not explicitly link chemical structures with their quantum chemical calculations, a preferred method in AI-assisted molecular design. Further experimental research is required to obtain the data of SMM behavior under uniform conditions.
However, this simplified approach has its advantages. It reduces the need for complex computational calculations and avoids the challenging task of simulating magnetism. Prof. Akitsu concludes: "Adopting such an approach can guide the design of innovative molecules, bringing about significant savings in time, resources, and costs in the development of functional materials."
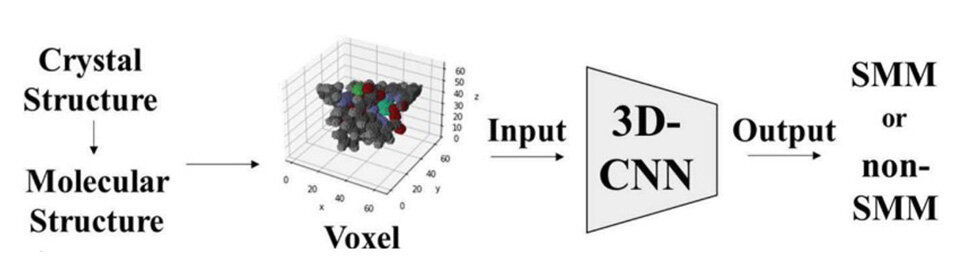
Image title: The steps involved in SMM prediction
Image caption: The novel method uses deep learning to identify single-molecule magnetic (SMM) materials based on their crystal structure alone.
Image link: https://doi.org/10.1107/S2052252524000770
Image credit: Takashiro Akitsu from Tokyo University of Science
License type: CC BY 4.0
Usage restrictions: Can be reused with appropriate attribution
Reference
| Title of original paper | : | The prediction of single-molecule magnet properties via deep learning |
| Journal | : | International Union of Crystallography Journal (IUCrJ) |
| DOI | : | 10.1107/S2052252524000770 |
About The Tokyo University of Science
Tokyo University of Science (TUS) is a well-known and respected university, and the largest science-specialized private research university in Japan, with four campuses in central Tokyo and its suburbs and in Hokkaido. Established in 1881, the university has continually contributed to Japan's development in science through inculcating the love for science in researchers, technicians, and educators.
With a mission of "Creating science and technology for the harmonious development of nature, human beings, and society," TUS has undertaken a wide range of research from basic to applied science. TUS has embraced a multidisciplinary approach to research and undertaken intensive study in some of today's most vital fields. TUS is a meritocracy where the best in science is recognized and nurtured. It is the only private university in Japan that has produced a Nobel Prize winner and the only private university in Asia to produce Nobel Prize winners within the natural sciences field.
■
Tokyo University of Science(About TUS)

About Professor Takashiro Akitsu from Tokyo University of Science
Takashiro Akitsu is a Professor at the Department of Chemistry, Faculty of Science, Tokyo University of Science (TUS), Japan. He graduated from Osaka University and obtained his Ph.D. in Physical and Inorganic Chemistry in 2000 and went on to study physical and bioinorganic chemistry at Stanford, before moving to TUS. He joined the TUS as a Junior Associate Professor in 2008 and became a Professor in 2016. He has published 250 articles and book chapters and served as an editorial board member in many international peer-reviewed journals. His current research areas involve the study of imines, Schiff bases, coordination chemistry, and crystal structures.
Laboratory website 
Official TUS website 

