2020.05.26 Tuesday
Modern Problems, Primitive Solutions: A Glimpse into Archaic Protein Synthesis Systems
Scientists show, for the first time, how protein synthesis in an ancient organism could have inspired the molecular tools used by modern organisms
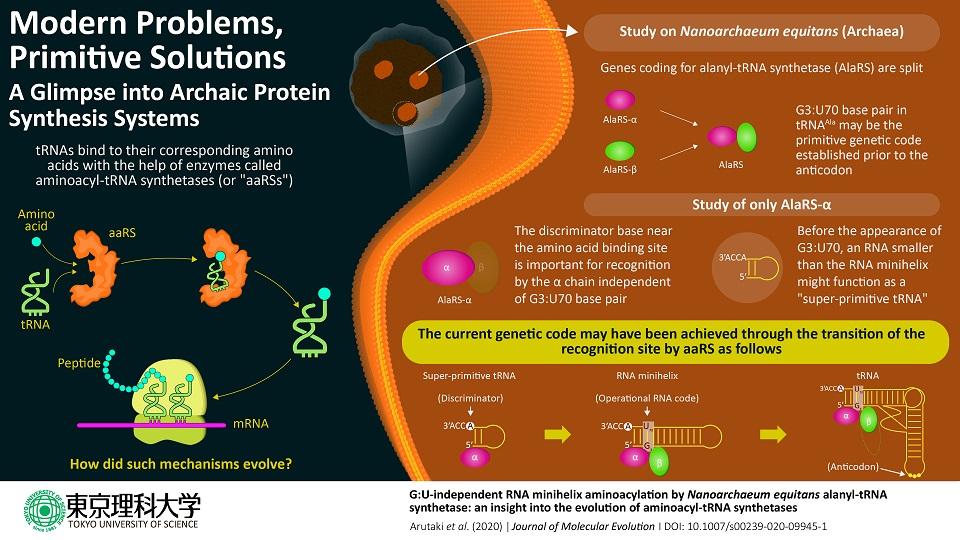
The interaction between 'transfer RNAs' and the enzymes that help them in protein synthesis has always been the key area of interest for understanding the evolution of the genetic code. Now, a team of scientists reports that a subunit of 'alanyl-tRNA synthetase' enzyme of the primitive microorganism Nanoarchaeum equitans can mimic the super-primitive tRNA 'aminoacylation' independent of the presence of a special 'G3:U70' base pair, a property previously unseen in primitive organisms.
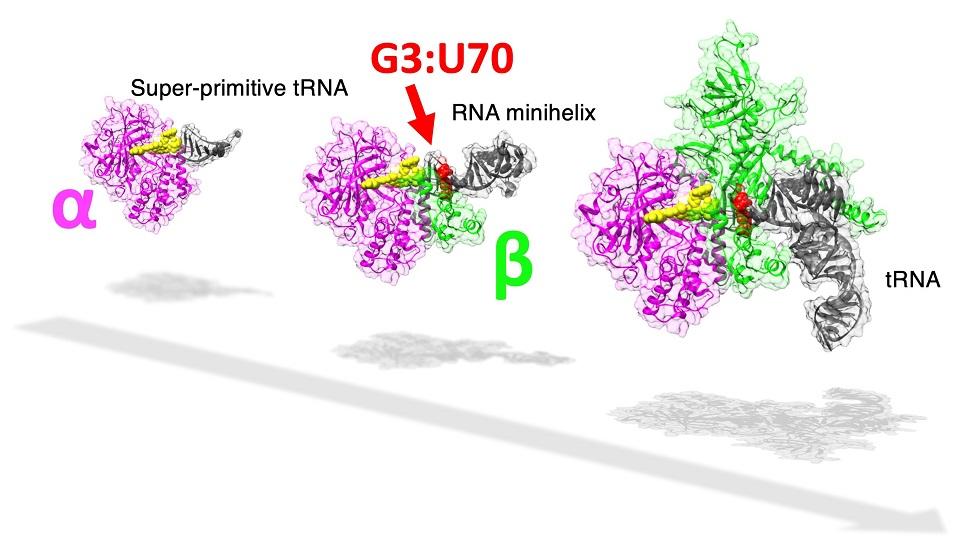 |
The study highlights the possible mechanisms of evolution of the current genetic code through the transition of the recognition site by aaRS. Image credit: Tadashi Ando (Tokyo University of Science)
In cells, protein is synthesized based on the genetic code. Each protein is coded by the triplet combination of chemicals called "nucleotides," and a continuous "reading" of any set of triplet codes will, after a multi-step process, result in the creation of a chain of amino acids, a protein. The genetic code is matched with the correct amino acid by a special functional RNA aptly named transfer RNA or tRNA (which, incidentally, is itself composed of its own type of "codes"). An enzyme called "aminoacyl-tRNA synthetase" or aaRS accurately assigns a specific amino acid to the correct "code" through a tRNA by recognizing unique structural components called 'identity elements' on the tRNA. In the case of the amino acid alanine, the identity element for recognition by the enzyme alanyl-tRNA synthetase (AlaRS) is an unlikely base pair "G3:U70," present in the minihelix structure (amino acid-accepting upper half region) of tRNA. Considering its importance in the recognition of the code, the base pair is popularly known as the "operational RNA code."
The evolution of this complex tRNA-aaRS system is a fascinating enigma, as the existing evolutionary evidence suggests that the upper half of the tRNA containing this operational code appeared earlier in evolutionary history than the lower half part that binds to the triplet code of mRNA. Interestingly, in a primitive microorganism, Nanoarchaeum equitans, the genes coding for each AlaRS subunit α and β are split, with the two genes being separated by half the length of the chromosome.
This interesting fact inspired a team of scientists at Tokyo University of Science, led by Prof. Koji Tamura, to hypothesize that these split forms of AlaRS in N. equitans might be connected with the evolutionary history of aaRS enzyme activity.
Prof. Tamura emphasizes the significance of their study, published in Journal of Molecular Evolution, in the evolutionary context, "AlaRS-α shows the G3:U70-independent addition of alanine to RNA minihelix regions. Our data indicate the existence of a simplified process of alanine addition to tRNA by AlaRS early in the evolutionary process, before the appearance of the G3:U70 base pair."
 |
The aforementioned minihelix parts of tRNAs were previously known to function as the region of occurrence of addition of amino acids by many aaRSs. To understand the interaction process of the minihelix (minihelixAla) of alanine-specific tRNA (tRNAAla) and AlaRS subunits, the researchers cloned the coding sequences of α and β subunits of N. equitans and then purified the synthesized proteins.
The researchers noticed that, at a relatively high concentration, AlaRS-α alone was capable of adding alanine to both tRNAAla and minihelixAla. Then also observed that AlaRS-α alone interacts with the end of the alanine-accepting region of tRNAAla, but not with the G3:U70 base pair. This was in stark contrast to prior knowledge regarding tRNAAla and AlaRS system. In brief, when both AlaRS-α and AlaRS-β were present, AlaRS behaved in a G3:U70-dependent manner, but working alone, AlaRS-α could add alanine to tRNAAla and minihelixAla in a G3:U70-independent manner. The researchers deduced that "the G3:U70 may be a late-arriving 'operational RNA code,' relevant to later alanylation systems incorporating further specificity through the evolution of the AlaRS-β subunit."
So, what makes the findings of this study so important? Prof. Tamura explains the significance of the striking results of their research, "our findings reveal for the first time that a G3:U70-independent mechanism of alanine addition exists. Furthermore, using 'RNA minihelix' molecules, which are considered to be the primitive form of tRNA, we could also illuminate the 'morphology' of tRNA before the evolutionary appearance of the G3:U70 base pair."
While discussing the broader implication of their study, Prof. Tamura comments thoughtfully "The breakthroughs in science almost always came from the curiosity-driven research, and the results of our study approach the mystery of the origin of life. It has the potential to transform many areas". His team is now focusing on an extensive structural analysis using the mutants of N. equitans AlaRS-α, but their current findings, published in August issue in printing and selected as the cover of the August issue, are enough to give cause to rethink chapters scientists that have believed to be fundamental in evolutionary history!
| Reference | ||
| Title of original paper | : | G:U-independent RNA minihelix aminoacylation by Nanoarchaeum equitans alanyl-tRNA synthetase: an insight into the evolution of aminoacyl-tRNA synthetases |
| Journal | : | Journal of Molecular Evolution |
| DOI | : | 10.1007/s00239-020-09945-1 |
The full paper, provided by Springer Nature SharedIt, can be accessed at https://rdcu.be/b32lR
About The Tokyo University of Science

Tokyo University of Science (TUS) is a well-known and respected university, and the largest science-specialized private research university in Japan, with four campuses in central Tokyo and its suburbs and in Hokkaido. Established in 1881, the university has continually contributed to Japan's development in science through inculcating the love for science in researchers, technicians, and educators.
With a mission of "Creating science and technology for the harmonious development of nature, human beings, and society", TUS has undertaken a wide range of research from basic to applied science. TUS has embraced a multidisciplinary approach to research and undertaken intensive study in some of today's most vital fields. TUS is a meritocracy where the best in science is recognized and nurtured. It is the only private university in Japan that has produced a Nobel Prize winner and the only private university in Asia to produce Nobel Prize winners within the natural sciences field.
About Professor Koji Tamura from Tokyo University of Science
Professor Koji Tamura is a Professor at the Department of Biological Science and Technology at the Tokyo University of Science. He is a renowned researcher with 14 years of experience in teaching. His research involves multidisciplinary study of the origin of life on Earth. His area of interest lies in the subjects like analysis of aminoacyl-tRNA synthetases mediated aminoacylation of tRNA and its relationship to the origin of the genetic code, peptide bond formation on the ribosome. He has more than 40 publications in prestigious international journals.
PROFILE
Tokyo University of Science, Faculty of Industrial Science and Technology, Department of Biological Science and Technology : https://www.tus.ac.jp/en/fac/p/index.php?4d37
Funding information
This research was supported by JSPS KAKENHI Grant Number JP17K19210.

